Fraud in food sales is a huge problem around the world, but such transactions can be classified as criminal activity where the hope of profit is high, but the risks and penalties are small for the individuals who engage in the occupation. In 2013, there was a large-scale misconduct in Europe, where horsemeat was sold on a large scale as beef in European supermarkets.
The 2013 horsemeat scandal caused a great deal of awareness among consumers and producers of beef in Europe. The case also opened the eyes of consumers to how complex and delicate the value chain of meat production really is. Many companies involved in this valuable branch of food production also realized the need for rapid, robust and cost-effective methods to verify the origin of products in the food market.
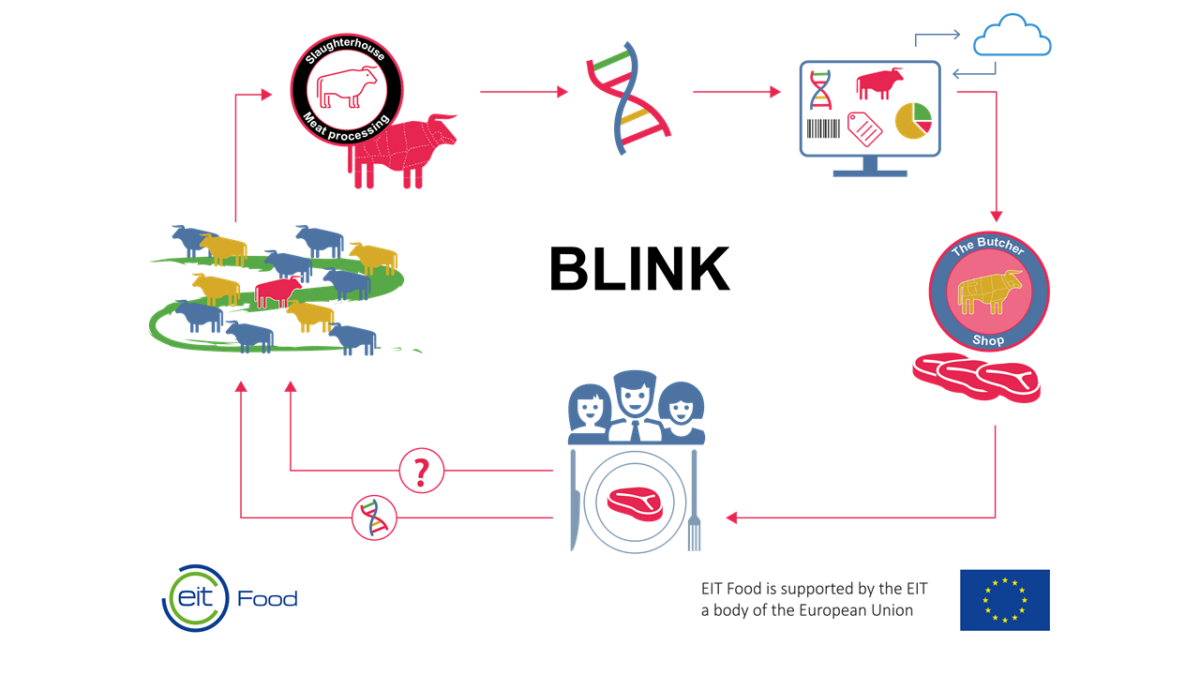
In recent years, Matís has led an international research project, fully funded by the European competition fund EIT Food. The project is called BLINK, but its goal is to develop a new traceability system for beef. The method is based on the great advances that have taken place in the last decade in genetic analysis technology and is comparable to methods that will be used in the selection of genomes in the Icelandic cow breed, which the aim is to implement in Iceland in the coming years.
The idea behind the methodology is that when slaughtering cattle, a biological sample, for example a hair sample or a tissue sample, is taken from the animal. The sample is then sent to a laboratory where the genetic material is isolated and thousands of genetic markers are analyzed in each and every grab. This genetic information is then used to create a unique "bar code" for each and every cattle that passes through a specific slaughterhouse. A genetic bar code has the advantage over traditional bar codes, which everyone knows from the supermarket, that it accompanies the meat wherever it goes and can not be changed in any way. The aforementioned genetic information, together with supporting documents, is finally placed in a database. If there is a suspicion that there is fraud somewhere in the chain from beef on a leg to a piece of meat on a plate, a sample of the meat can be sent for genetic analysis and it can be determined whether the origin of the beef is the same as the food packaging indicates.

This methodology could easily be applied to verify the origin of Icelandic beef from supermarkets or restaurants. Recent research has confirmed that the Icelandic cow population is genetically very different from other cattle. With a simple genetic test, it would be possible to confirm the origin of Icelandic beef.
The project will end in January next year. Following this, Matís' foreign partners will draw attention to the methodology among European meat producers. It is hoped that the technology will be able to be used in general in the western world in the next few years and that it can also be used for traceability of meat products of other livestock species.